Chapter 8 and Chapter 20 Ana Luis ana.luis@medkem.gu.se 2025/04/28
Protein Biosynthesis
(Translation) Chapter 8 and Chapter 20 Ana Luis ana.luis@medkem.gu.se 2025/04/28
Learning goals
• Explain how nucleic acid information is translated into an amino acid sequence and define the role of aminoacyl tRNA synthetases in this process • Protein biosynthesis requires the translation of nucleotide sequences into amino acid sequences • Describe features of the genetic code • tRNA structure and function • Aminoacyl-tRNA synthetases establish the genetic code
• Define the role of ribosomes in protein synthesis • Structure and function of ribosomes • Mechanism of protein synthesis (initiation, elongation, translocation and termination)
• Describe how certain chemicals can inhibit protein synthesis • Antibiotics and toxins inhibit protein synthesis
Why is important to understand the translation mechanisms?
TRANSLATION
Inhibition of translation of specific mRNAs can lead to diseasess
- Fragile X mental retardation syndrome:
- absence of the set of protein isoforms, derived from alternative splicing of the Fragile X mental retardation gene 1 (FMR1)
Bacterial toxins can block of protein biosynthesis protein biosynthesis
- Diphtheria toxin
Several antibiotics are inhibitors
- Streptomycin
- Tetracycline
- …..
⸻
Translation = the process of protein biosynthesis
Adenine (A) Cytosine (C) Guanine (G) Uracil (U) Nucleic acid
→
Amino acids (A) Alanine (R) Arginine (N) Asparagine (D) Aspartic acid (C) Cysteine (E) Glutamic acid (Q) Glutamine (G) Glycine (H) Histidine (I) Isoleucine (L) Leucine (K) Lysine (M) Methionine (F) Phenylalanine (P) Proline (S) Serine (T) Threonine (W) Tryptophan (Y) Tyrosine (V) Valine
Translation = the process of protein biosynthesis
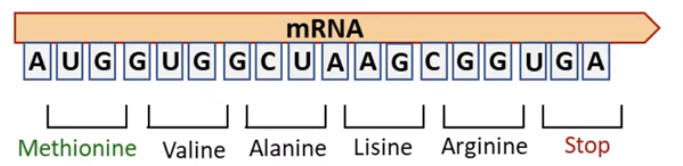
The basics of protein biosynthesis are the same across all kingdoms of life: • mRNA is decoded in the 5′-to-3′ direction one codon at a time • the protein is synthesized in the amino-to-carboxyl direction mRNA A U G G U G G C U A A G C G G U G A Methionine Valine Alanine Lisine Arginine Stop 5’ 3’ Amino group Carboxyl group Amino group Carboxyl
⸻
tRNA
a.a. Translation = the process of protein biosynthesis • anticodon - portion of the tRNA that base pairs with the codon C G I 3’ 5’ G C C 5’ 3’ Codon • codon - three coding bases on the mRNA template • transfer RNA (tRNA) - function as adaptor molecules between a codon and an amino acid (a.a.) Anticodon mRNA The basics of protein biosynthesis are the same across all kingdoms of life: The correct protein biosynthesis requires an accurate recognition of codons by anticodons
General characteristics of Transfer RNA (tRNA) molecules
• Each tRNA is a single chain containing between 73 and 93 nucleotides. • The secondary structure resembles a cloverleaf (~50% of the nucleotides are base-paired) Five groups of bases are not base-paired, but participate in hydrogen-bonding interactions: • 3′ CCA terminal region (acceptor stem) • TψC loop • “extra arm” • anticodon loop • DHU loop • The three-dimensional structure is L-shaped • tRNAs contain 7 to 15 unusual bases • methylated or demethylated derivatives of A, U, C, and G
⸻
• At the 3’ end, an activated amino acid is attached to a hydroxyl group of adenosine in the CCA region of the acceptor stem • the CCA region has the ability to change its conformation during protein synthesis • The anticodon loop is near the center of the sequence General characteristics of tRNA molecules
⸻
Genetic code
C G I 3’ 5’ G C C 5’ 3’ Codon Anticodon mRNA • Genetic code: the relation between the sequence of bases in DNA and the sequence of amino acids in proteins mRNA A U G G U G G C U A A G C G G U G A Methionine Valine Alanine Lysine Arginine Stop 5’ 3’ DNA PROTEIN
⸻
Amino acids are encoded by groups of three bases starting from a fixed point
• Features of the Genetic code: – three nucleotides (codon) encode an amino acid – has directionality – nonoverlapping – has no punctuation – is degenerate (most amino acids are encoded by more than one codon) mRNA A U G G U G G C U A A G C G G U G A Methionine Valine Alanine Lysine Arginine Stop 5’ 3’ • 61 codons encode specify amino acids (20 in total) • 3 codons are stop codons that designate termination of translation.
⸻
’Wobble’ effect in base-pairing
’Wobble’ effect: Some tRNA molecules can recognize more than one codon tRNA a.a. 1 C G I 3’ 5’ G C C 5’ 3’ Codon Anticodon mRNA tRNA a.a. 1 C G I 3’ 5’ G C A 5’ 3’ Codon Anticodon mRNA
⸻
’Wobble’ effect in base-pairing
Anticodons base-pair with codons ’Wobble’ effect: Some tRNA molecules can recognize more than one codon Codons that differ in either of their first two bases (from 5’) must be recognized by different tRNAs. The first base of the anticodon (5’) determines the degree of wobble • The redundancy, or degeneracy, of the genetic code indicates that recognition of the third base of a codon is sometimes less discriminating than the other two “wobble” = steric freedom in the pairing of the first base of the anticodon with the third base of the codon
⸻
’Wobble’ effect in base-pairing
“wobble hypothesis”: established hypothesis that predicts the binding of anticodons to codons TABLE 30.2 Allowed pairings at the third base of the codon according to the wobble hypothesis Anticodons base-pair with codons C C G
⸻
’Wobble’ effect in base-pairing
“wobble hypothesis”: established hypothesis that predicts the binding of anticodons to codons TABLE 30.2 Allowed pairings at the third base of the codon according to the wobble hypothesis C U A/ G
⸻
’Wobble’ effect in base-pairing
“wobble hypothesis”: established hypothesis that predicts the binding of anticodons to codons TABLE 30.2 Allowed pairings at the third base of the codon Example: If first base of the anticodon is inosine, the anticodon can recognize three different codons Inosine is formed by the deamination of adenosine • has a heterocyclic nitrogen base that can form hydrogen bonds with adenine, cytosine and uracil The purine base inosine pairs with cytidine, uridine or adenosine
⸻
Amino acids required for protein biosynthesis must first be attached to specific tRNA molecules
• The attachment of a given amino acid to a particular tRNA establishes the genetic code Aminoacyl-tRNA synthetases Aminoacyl-tRNA synthetases attach specific amino acids to tRNAs tRNA amino acid Aminoacyl-tRNA amino • acid
⸻
Ester linkages couple amino acids to tRNA
The process of attaching an amino acid to tRNA is called aminosylation CCA arm of tRNA • Amino acids are bound to the 3’end of the tRNA via an ester bond between the carboxyl group on the amino acid and either the 2’ or 3’ hydroxyl group of the terminal adenosine of the tRNA 3’ Aminoacyl-tRNA: Amino acid bound to tRNA 3’
⸻
Each aminoacyl-tRNA synthetase is specific for a given amino acid
How aminoacyl-tRNA synthetases evolve to differentiate between different amino acids? A closer look at the amino acid threonine, valine and serine
⸻
Threonyl-tRNA Synthetase contains an activation site
Each aminoacyl-tRNA synthetase is specific for a given amino acid How aminoacyl-tRNA synthetases evolve to differentiate Threonine, Valine and Serine? Activation site responsible for activating threonine by binding it to adenosine triphospate (ATP) and further transfer of these amino acid to the tRNA molecule • Aminoacyl-tRNA synthetases have highly discriminating amino acid activation sites
⸻
Each aminoacyl-tRNA synthetase is specific for a given amino acid
• To avoid coupling to the incorrect amino acid, threonyl-tRNA synthetase (tRNAThr) contains a zinc ion at the active site that binds to the amino and hydroxyl groups of threonine • Valine is similar in overall structure to threonine but lacks the hydroxyl group, so it does not bind to tRNAThr • Serine is occasionally linked to tRNAThr because of the presence of the hydroxyl group (Thr) (Val) (Ser) How aminoacyl-tRNA synthetases evolve to differentiate Threonine, Valine and Serine?
⸻
Threonyl-tRNA Synthetase contains an activation site and an editing site
Activation site responsible for activating threonine by binding it to adenosine triphospate (ATP) and further transfer of these amino acid to the tRNA molecule Editing site: acts as a profreader and removes any incorrect bound amino acid from the tRNA molecule
⸻
Proofreading by Aminoacyl-tRNA Synthetases Increases the Fidelity of Protein Biosynthesis
• Threonyl-tRNA synthetase has an editing site that hydrolyzes Serine if this is linked to threonine-tRNA • Because Thr contains an extra methyl group, it is sterically excluded from the editing site • The aminoacylated CCA arm of the tRNA is flexible and can swing out of the activation site and into the editing site to remove Ser • Most aminoacyl-tRNA synthetases contain editing sites and activation sites to ensure very high fidelity. Threonyl-tRNA synthetase Thr-tRNA
⸻
Aminoacyl-tRNA synthetases interaction with tRNA
Threonine-tRNA synthetase tRNA • Threonine-tRNA synthetases bind to both the acceptor stem and the anticodon loop of the tRNA Aminoacyl-tRNA synthetases assign a particular amino acid to a specific tRNA - the true translators of the genetic code • Some synthetases recognize their tRNA partners primarily on the basis of their anticodons • Synthetases may also recognize other aspects of tRNA structure that vary among different tRNAs • many of the recognition sites are loops rich in unusual bases Number of interactions between tRNA and aminoacyl-tRNA synsthetases
⸻
The ribosome is the site of protein synthesis
Ribosomes coordinate the interplay of aminoacyl-tRNAs, mRNA, and proteins aminoacyl-tRNAs
⸻
The E. coli ribosome has a sedimentation coefficient of 70S
composed of: • large (50S) subunit • small (30S) subunit 34 proteins (L1–L34) 23S rRNA 5S rRNA 21 proteins (S1–S21) 16S rRNA • Two-thirds of the mass of ribosomes is RNA • Ribosomal RNA (rRNA) is the catalyst for protein synthesis
⸻
The ribosome has three binding sites for transfer RNAs
• Three tRNA-binding sites: • A site (aminoacyl) • P site (peptidyl) • E site (exit) • mRNA fragment is bound within the 30S subunit • Each tRNA contacts both 30S and 50S subunits
⸻
Overview of mechanism of protein biosynthesis
Initiation, Elongation, Translocation and Termination Initiation
⸻
Initiation of translation
• Each initiator region usually displays: • start codon: AUG (Met) or sometimes GUG, rarely UUG • purine-rich sequence ~10 nt upstream • Bacterial mRNAs often encode several polypeptides • Shine–Dalgarno sequence = purine-rich region binding rRNA to position initiator codon in P site
⸻
Bacterial protein synthesis is initiated by N-formylmethionyl-transfer RNA
Steps: 1. Met linked to tRNAfMet by aminoacyl-tRNA synthetase 2. Met amino group is formylated 3. fMet-tRNAfMet placed in P site (note: tRNAMet inserts internal Met)
⸻
Initiation factors (IF1, IF2, IF3) 1. IF1 + IF3 bind 30S to prevent premature binding to 50S 2. IF2(GTP) + fMet-tRNAfMet + mRNA bind to 30S → 30S initiation complex 3. Structural changes release IF1 + IF3 4. IF2 stimulates 50S binding + GTP hydrolysis → 70S initiation complex Streptomycin binds 30S and blocks fMet-tRNAfMet binding (bacteria)
⸻
Elongation
Elongation factors deliver aminoacyl-tRNAs to the ribosome
⸻
Elongation factors deliver aminoacyl-tRNAs
Steps: 1. EF-Tu-GTP binds aminoacyl-tRNA → delivers to A site 2. Correct codon recognition → GTP hydrolysis → EF-Tu-GDP leaves 3. EF-Ts binds EF-Tu-GDP 4. EF-Ts releases GDP → GTP binds → EF-Tu-GTP regenerated Tetracycline binds 30S → blocks aminoacyl-tRNA binding
⸻
Elongation – peptidyl transferase
• Amino group of A-site tRNA attacks carbonyl of P-site peptidyl-tRNA • Peptidyl transferase center on 50S catalyzes peptide bond formation • Reaction is thermodynamically spontaneous but accelerated by ribosome positioning/orientation
⸻
Translocation
Translocation repositions tRNAs and mRNA EF-G (translocase) catalyzes 1-codon movement (requires GTP) 1. EF-G-GTP binds near A site 2. GTP hydrolysis → conformational change → peptidyl-tRNA shifts A→P After peptide bond formation: chain is in P site (50S), anticodon still in A site (30S)
⸻
Termination
Release factors (RFs): • RF1 + RF2 recognize stop codons (UAA, UGA, UAG) • RF3 (GTPase) removes RF1/RF2 RF1/RF2 bind stop codon in A site RFs contain conserved GGQ motif with methylated Gln → promotes hydrolysis of ester linkage → releases polypeptide
⸻
Bacteria: transcription and translation are coupled
Minimal time gap between transcription and translation Polysome: multiple ribosomes translating one mRNA simultaneously
⸻
Difference between bacterial and eukaryotic protein biosynthesis
Bacteria 70S: 50S+30S Eukaryotes 80S: 60S+40S Initiation differences: • Initiating amino acid = Met (not fMet) • No Shine–Dalgarno • Start site usually first AUG from 5’ end • Many more eIFs Eukaryotic mRNA circularization: 5’-cap proteins + PABP at 3’-poly(A) interact → circular mRNA
⸻
TABLE 30.4 Antibiotic inhibitors of protein biosynthesis
Streptomycin/aminoglycosides — inhibit initiation + misreading (bacteria) Tetracycline — blocks aminoacyl-tRNA binding (30S) Chloramphenicol — inhibits peptidyl transferase (50S) Cycloheximide — inhibits translocation (eukaryotes) Erythromycin — blocks translocation (50S) Puromycin — premature termination (aa-tRNA analog)
⸻
Diphtheria toxin
Blocks protein biosynthesis in eukaryotes by inhibiting translocation
⸻
Concepts
DNA RNA Protein Translation Transcription Messenger RNA (mRNA) Initiation factors Elongation factors Release factors Aminoacyl tRNA synthetases Transfer RNA (tRNA) Ribosome (ribosomal RNA + proteins) EF1 EF2 EF3 EF-Tu EF-Ts EF-G RF1 RF2 RF3 • Genetic code • Degeneracy • Codon • Anticodon • Transfer RNA • ’Wobble’ effect • Aminoacyl-tRNA synthetases • Aminoacyl-tRNA • Aminosylation • Aminoacyl-tRNA synthetases activation site • Aminoacyl-tRNA synthetases editing site • Ribosome • Ribosomal RNA • Ribosome E site, P site and A site • Shine-Dalgarno sequence • Initiation, elongation, translocation, termination • Initiation factors • Elongation factors • Termination factors • Polysome • Inhibition of trancription